In my last entry I demonstrated the difficulties of determining the half-relationships after receiving the DNA results of my half-first cousin. Within an endogamous group, that could be even more difficult as we see larger amounts of DNA shared.
While the ISOGG Wiki Autosomal DNA Statistic page can list the average amount of centimorgans shared, Blaine Bettinger’s The Shared cM Project demonstrated that the minimum and maximum amounts shared can vary. This becomes more evident as the distance of relationship increases.
Within an endogamous group it makes sense that having more than one pair of common ancestors may increase that amount. The same would apply if you descend from the same common ancestor multiple times. Both would produce higher amounts shared.
A few months ago I got the results of my aunt believed to be a full-sister of my mother. My aunt suspected that her father was not her biological father. And she was right. But she was not the only one who knew of this, but the rest of the family, particularly the ones of my generations believed that this Aunt’s father was her biological father and did not suspect otherwise.
From my mother’s Family Finder (autosomal) match list at FTDNA:
The top is my mother’s sister while the one right below it belongs to my half-1st cousin whose father George was mentioned in the last entry – Determining half-relationships with Polynesians.
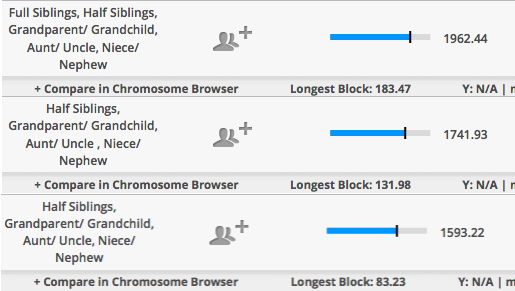
Initially I was confused by the total amount since I knew it was more than what I shared with two of my half-brothers. This is how two of my half-brothers compare to me and to each other.
So my mother and her sister did share a bit on the high-end for half-siblings, but low end for full-siblings. These are the predicted averages shared for siblings vs. half-siblings.
The next step was to take a look at the X chromosome. For half-sisters who had the same father, they would share an entire X chromosome based on how the X is inherited. To my surprise, it looked like someone took a razor blade and sliced out some pieces of the image.
For half-sisters they share a lot compared to what I saw when comparing my half-brothers to each and to me. Also, I decided to include both the default 5+cM setting and the 1+cM. With my brothers, we hardly get anything when I lower it to 1+cM. But with my mother and aunt, you can see a difference although chromosomes 4 and 18 are more likely to be IBS, but given the situation (endogamy, small communities, & isolation) it just may be IBD from a very long time ago.
So the X was not helping me one bit since I thought maybe they were areas on the chromosome that could not be read – no calls.
I immediately uploaded to GEDmatch for further analysis. No surprise that when I looked at the X, it was the same exact thing. Knowing that it wouldn’t be helpful, I turned to the other 22 pairs of chromosomes.
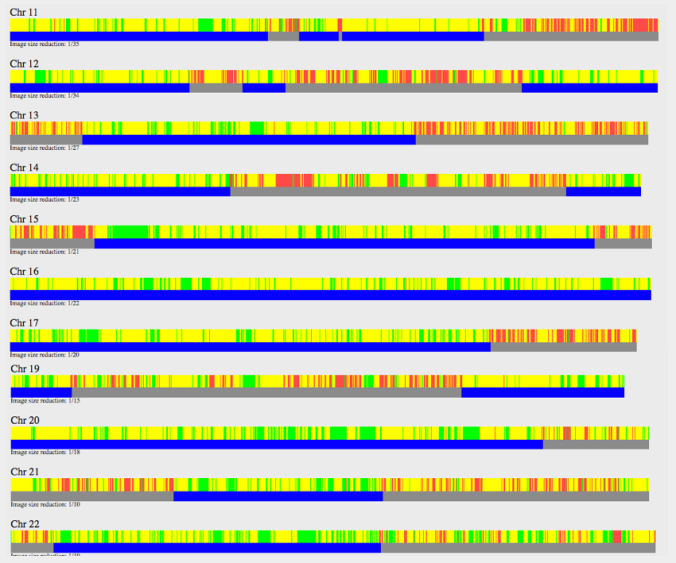
What you would be looking for in full-siblings are full-identical regions (FIR) which are the green sections on the bar graph. Here is an example of my 1st cousins, a brother and sister.
About 25% will be fully identical. You can read more about how much full versus half-identical regions siblings would share at ISOGG’s Wiki – Fully Identical Region page.
This is what my mother and aunt showed.
There are only small chunks of FIR rather than long segments of it that you would see in full-siblings. So this confirms a half-sibling relationship.








Hi Kalani-
I am of mainly Northern European descent. However on all of the Calculators at GEDmatch, I get 0.7%-1.4% Melanesian/Papuan/Oceanian. I don’t have any Pacific Islander ancestors, and I know this is a very distinct population. What are your thoughts on this?
J bower
LikeLike
Hi Jacob. In most cases anything less than 2% is considered statistical noise. But for some people, that noise may be valid only because they know of a history. So if you know you do not have any history of Oceanic ancestors, then it is noise for certain. And it depends on which admixture calculator you use. If I use the EthioHelix calculator intended for those of African ancestry and specifically select EthioHelix K10 + Japanese, I come up with the following results:
Nilo-Saharan 2.10
Omotic 1.39
Khoi-San 0.40
Hadza 3.24
Biaka-Pygmy 1.72
North-Africa 26.02
Japanese 65.14
Does that mean I am of those ancestries? More so, does that mean I am 26% North-African and 65% Japanese? Our ancestors did not come from Japan and I know for a fact where my ancestors from at least the mid-1700s for my paternal side and for the 1600s on my maternal side were from. They were not from either North Africa nor Japan. These admixture calculators have various categories based on samplings of specific people and now they are trying to compare your genome to these specific samples only.
LikeLike
Aloha, I have not taken any DNA yet but I was interested in the X chromosome. There is a Haplogroup X which is divided into 2 clades: Haplogroup X1 and X2.
X1 origin is of Northern Africa.
X2 origin is of Eurasian descent namely Syria, Lebanon, Israel and a small community of Jordan, and the Native American Indians near the great lakes. The X chromosome that you may rule out may be something you may want to investigate more. Because the X chromosome can only derive from X1 or X2 of Haplogroup X. I believe that your mother’s chromosome X is of X2. You may need to investigate to verify Haplgroup X2.
This group is an older European group than those of present day European groups that arrived during Captain Cook to present day Hawaiians/Native Hawaiians.
For more conclusive studies on Haplogroup X2. Contact person Rod Meldrum 801-429-9751. He will be able to help you. Thank you for sharing your families DNA. Especially the X chromosome.
Mahalo nui loa. brada Paule
LikeLike
Aloha e Paul,
What you are referring to are the haplogroup for mtDNA, not the X chromosome.
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1180497/
Our mitochondrial (mtDNA) haplogroup is of the Polynesian motif – B4a1a1. We are actually B4a1a1c, a subgroup of the Polynesian motif. Many Polynesians are either the following:
B4a1a1
B4a1a1a
B4a1a1a1
B4a1a1a3 (now this is B4a1a1c, which is what I am)
The X chromosome is a sex chromosome (like the Y chromosome) and is a part of our autosomes, or 23 pairs of the autosomes. The haplogroups are applied only to Y-DNA and the mitochondrial DNA.
LikeLike
Aloha Kalani, yes I am referring to mtDNA variant markers and not sex thank you for correcting me. I’m interested in the origin of our Hawaiian race. Would your mtDNA have these variants 16213A and 200G. That this variants orginate from Haplogroup X2. I am considering taking my own DNA to search for these variants. I took interest in your X chromosome. To see DNA result of Hawaiian descendants if these markers were present in your mtDNA. I have mistaken chromosome for variant markers. What type of DNA testing would you consider (me) to find these variant markers. I’m interested in our origin. Not so much the motif for now although it is equal important.
LikeLike
Here’s the current Phylotree which is used.
http://phylotree.org/
Specifically I am of haplogroup B which comes out of East Asia.
http://phylotree.org/tree/B.htm
According to that link, our B4a1a1c line has 2 defining markers: C1185T G4769A
As you mentioned earlier, X is a very old line. I remember reading in one of the research that the Polynesian motif is dated to be older than 7,000 yrs. The X is much, much, much older than that.
And since you seem to be more into the markers present in haplogroup, familytreedna.com is the best to get mtDNA tested and Y-DNA tested. I’m not too savvy on the Y-DNA. And a few of the haplogroup projects are working with a few members of my Polynesian DNA project to get a more thorough testing done in order to identify some of these not-so-known markers since we do not have a lot of testees of oceanic origin.
You can see results of the Y and mtDNA of group members here:
https://www.familytreedna.com/groups/hawaiianancestry/dna-results
But for the mtDNA, you are seeing mostly the defining makers on HVR1 and HVR2. Some people have left their coding region private, some people make their results not seen in these projects except by the project administrator like myself. But there (above link) is where you can see our results. My kit is 285846.
LikeLike
I liked about the coding region. It’s not shown in the public forum. 🙂
https://www.familytreedna.com/public/hawaiianancestry?iframe=mtresults
LikeLike