In my last blog entry AutoCluster Endogamy tool at GEDmatch.com (Part 1), I covered briefly about the settings that are adjustable when ready to produce clusters and what is suggested for Polynesians. I also mentioned Leah Larkin referring to different levels or degrees of endogamy based on the average size segment and given that size, what works best or how to approach your DNA matches. I had to play around with it quite a bit in order to get decent clusters.
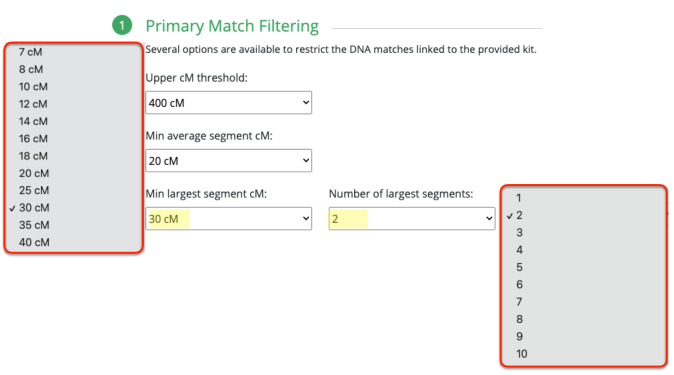
First, understanding the high settings put in place when you select “Highly Endogamous.”
So the minimum largest segment is 30cM, which is what I’ve been promoting for nearly 11 years. This was based on my own observation when I tested a second 1C1R and could compare that 1C1R with another 1C1R who had tested a few months prior, and are 2C to each other. Given that we have a lot of predicted 2nd to 3rd cousin matches (100cM – 300cM) where the largest segment size rarely would exceed 20cM. With these two 1C1R who are 2C to each other, I noticed that their largest segment was 41cM.
It wouldn’t be till about 2 or 3 years later when I heard others somewhat following that same analogy and seeing the significance of a largest segment size indicating a closer ancestral connection versus an endogamous one. This was specific with Ashkenazi Jewish background and how they seem to be set on 20cM. By this time, I had already determined that 30cM would be best. Also, with other Polynesians who share a true 2nd to 3rd cousin relationship, their largest segment would be larger than 20cM. And among all the endogamous matches, it rarely would exceed 20cM.
So that was the reason why I determined that 30cM was a good amount to be used in the Min largest segment cM. And while this blog entry is specific for this new AutoClustering tool for endogamy at GEDmatch, I have noticed that at MyHeritage, even with the endogamous matches that the largest segment size could exceed 30cM. However, what would also be indicative of an endogamous match vs. a truly close 2nd to 3rd cousin match, is the number of segments.
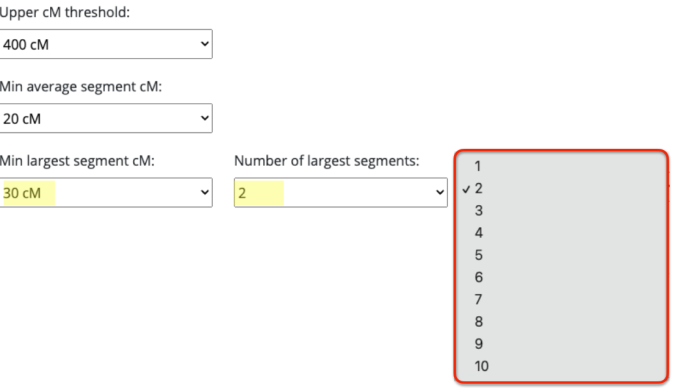
Taking a closer look and right next to the minimum largest segment size is the number of largest segments. Thought this was interesting and not sure if it’s necessary or not.
I am assuming that when it asks for minimum largest segment followed by the number of largest segments, that would mean it will have your smallest — largest segment size set at whatever number you have selected, times whatever you selected under number of largest segments. In other words, if you select 100cM min largest segment size, it will require that the smallest size you have is not smaller than 100cM. And the number of largest segments, say I select 10, it would require that you have at least 10 segments no smaller than 100cM.
You rarely would get a largest segment of the same size, or at least not that I have seen in both endogamous and non-endogamous matches. After all, these direct to consumer DNA testing companies are showing you the size of the largest segment that you have among all of the matching segments that you share. This is probably why I initially was not generating any matches/clusters simply because I had it set to 2. So my suggestion is to change it to 1.
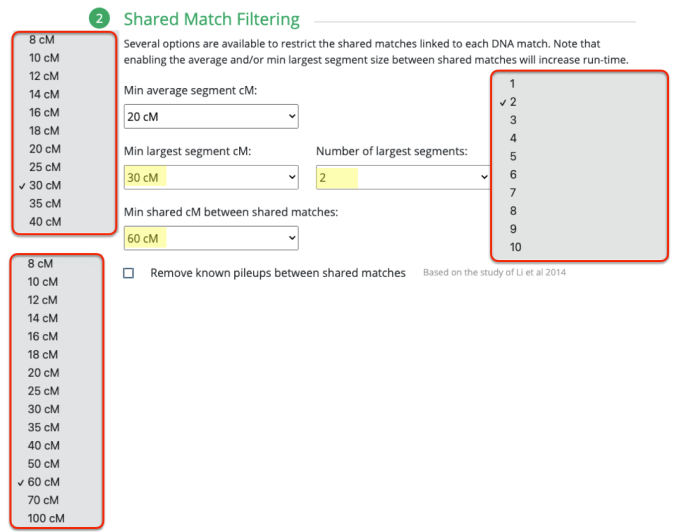
So all of those parameters are allowed under the Primary Match Filtering section. Then you have the Shared Match Filtering section which is nearly identical to the Primary Match Filtering section except you also have a minimum shared cM between shared matches which is what you also see when you run an autocluster with MyHeritage or Genetic Affairs directly.
With this parameter, you can tell it how much your DNA matches must share with each other to be considered to be put into a cluster. And what I did was set it to as low as 100cM since I have hundreds of matches from 100cM up to 200cM. My advice is that if you’re not admixed, or rather you have less foreign branches, definitely increase that higher than 100cM. It was easier for me to guess the numbers to use since I know how many matches I have, and out of these varying ranges of shared DNA, how many matches I would have for each.
For example, I have hundreds of matches predicted to be 2nd cousins (Ancestry). That is I have hundreds of matches sharing as low as 200cM and as high as 649cM. In the predicted 3rd cousin range I have over a thousand of these type of matches which range from 90cM to 199cM. And predicted 4th cousins, more than 24,000. These range as low as 20cM and as high as 89cM.
In my previous post I showed an example of what my autocluster looked like from MyHeritage and that I sorted it (by total shared cM) from the lowest to the highest. The lowest was 108cM, and from there it slowly went up. I had 12 matches sharing 108cM. I also had 12 matches sharing 109cM. The number of matches sharing about the same amount can be a lot. So understanding this will help you decide the best numbers or amounts to use when creating your clusters.
I am hoping that others from various endogamous groups start utilizing this new tool and am really curious how it will affect their research, expecting it to be for the better! Since I am still trying to generate various clusters by constantly adding in varying numbers, I will not be posting any examples of what they look like. Perhaps in a future blog post I will.
I also noticed that with a list of files when generating these autoclusters at GEDmatch, you also get csv files to be used in Gephi. I posted samples of that in my post from December 2022 called In-common-with, shared matches, and clustering. I will have to take time to also try to use these actual clusters and look to see how Gephi renders it.
Thanks Kalani for trying this out with your very complicated endogamous ancestry. I was looking forward to see a verdict, how well this new solution has specifically improved your situation. I haven’t found a final verdict and I’m not sure if you will write a blog post about it in the future but I would very much look forward to such or at least maybe hear your pros and cons in an answer to my comment.
LikeLiked by 1 person
I was happy that it actually did produce clusterS. That is more than 2 clusters! But the names didn’t look familiar, but understandable since I do not use GEDmatch anymore. I would really have to spend time looking into the DNA matches and try to see if there are any gedcoms attached, and if not, would have to research their lines, if not contact them. But I did notice that the clusters consisted of pretty distant matches. But again, I would really have to take time to look into it, because maybe I am wrong and they aren’t as distant as I thought.
LikeLike