HOW WE CONNECT
Now that I had figured out who my mother’s biological parents were it has become easier to find connections. (You can read about it here: https://hawaiiandna.wordpress.com/2015/08/01/finding-a-dna-connection-despite-endogamy/)
While there is one branch where I find a lot of relatives on my great-grandmother Rose Holbron’s side, I am slowly finding distant connections on my great-grandfather Frank Kanae’s side. Frank Kanae was Rose Holbron’s husband.
Earlier this week I received an email from a woman named Raychelle who saw me and my numerous kits of family members that I manage on GEDmatch.com as a match to her. I began the normal response, almost ready to dismiss her since many of these matches appear to be close when in reality we are usually distant, and for others, much more distant. And from what I could see, it wasn’t such a huge amount. At GEDmatch, Raychelle and my mother shares 62.9cM total, with a large segment of 10.7cM. So at least a 4th cousin level.
After I told her that she could find me on Ancestry (since she uploaded to GEDmatch via Ancestry) and look at my HOLBRON family tree, she found out that we have the LEWIS connection.

She is a 5th cousin to my mother, and a 5th cousin once removed (5C1R) to me. I come from Isaac Lewis who was known as Isaac Lewis Kanae or Isaac Kanae Lewis, and also known by the Hawaiianized version – Aikake Lui. While Raychelle comes from John George Lewis, and his Hawaiianized name was Keo Lui. My assumption is that Keo was short for Keoki (George). Keo could also be short for Keoni (John) and then there was the catholic version – Ioane for John.
But what was interesting is that she had this genealogy and I had updated mine from this to reflect what a couple of people have been researching.
According to the information that has been circulating at various sites on the internet, Isaac’s father – Captain Isaiah Lewis was the son of Captain Ezra Lewis. And John G. Lewis was the son of Captain John Lewis, who was Captain Ezra Lewis’ son but through a different wife. I listed them as spouse #1 and spouse #2 because different sites and people will switch the spouses showing Isaac as the son of one spouse, and another will show Isaac as the son of the other spouse, and vice versa for John G. Lewis.

So the question is, were Isaac and John full brothers, or (maternal) half-brothers? And if they were (maternal) half-brothers, were their fathers paternal half-brothers?
While all of this information going back that far is based solely on people creating these trees without further documentation, for now I am only going by what was documented. The trees habitually say that Polly was known as Sarah Pauline “Polly” Holmes. While I can understand that Polly could be a diminutive for Paula and Mary, I’m not so sure that these are the same person, especially since a lot of the information lists this Sarah Pauline “Polly” Holmes having been born in Massachusetts and died there, and that her husband Captain Isaac Lewis from Massachusetts too.
What we know for a fact according to testimonies from people who lived during the time of Polly Holmes and her father Oliver Holmes.

I am still in the process of confirming and documenting all of these ancestors, so for now I am considering Raychelle and I 5C1R, and that her 3x great-grandfather John George Lewis (Keo Lui) and my 4x great-grandfather Isaac Lewis Kanae (Aikake Lui) were full-brothers.
SHARED DNA SEGMENTS & CHROMOSOME MAPPING
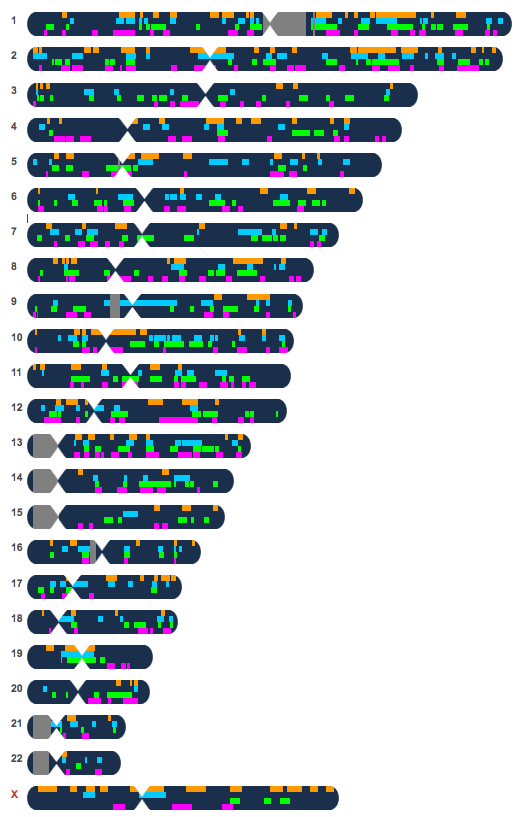
I compared Raychelle to all of the relatives to see which segments we all had in common. Any common segments or segments that multiple relatives share would indicate that segment was inherited from a common ancestor. In this case, Polly Holmes and her husband Isaiah Lewis.
And while autosomal DNA inherited from our common ancestor can remain in our genome for about 5 – 6 generations, there are some cases where it can span several generations and for some as we have seen, in larger segments. These larger segments tend to be passed on within generations entirely intact and having not recombined.
With endogamy, that may confuse things as it isn’t guaranteed that the shared segment came from that same common ancestor. Especially for Polynesians where we share many small segments. And these multiple segments may not be in common with other relatives, or rather these segments may not overlap as what I am about to demonstrate. So when looking to map out these segments, and at the 4x great-grandparent level, if the segments are really small, that may be suspect to being segments randomly inherited. It may or may not be from the common ancestor, or may come from the same common ancestor multiple times through their different descendants.
I first compared my brother Kaimi and Raychelle and looked for the chromosomes that should match my mom. Kaimi and I have different fathers, so I decided to use his to compare because his father is also Hawaiian.
I use Kaimi’s unphased and phased data to be sure that if there are extra segments that does not match our mother, then the presumption is that the segment came from Kaimi’s father. These were the results.

You can easily see how with the phased data the size of the segment is somewhat smaller if it doesn’t remain the same or disappear altogether.
The real work comes in when I compare Raychelle to my mom’s brother’s son Chris, her half-brother’s daughter Lena and her maternal half-sister Aunty Stella. The detailed specification of their relationship is to help you understand how they are related and know what is to be expected as far as sharing DNA with different relationships go.
What I did first was compare Raychelle to all of those family members mentioned and then see which of those matching segments actually matches up with what my mother matches. Here’s a diagram of how we are related and descend from Isaiah Lewis and Polly Holmes.

I’ll start first with Chris, the son of my mother’s brother Joseph.

While there were other segments that Raychelle shared with Chris, I am only comparing overlapping segments that are shared with my mom. There are 3 chromosomes where they share overlapping segments. Ch 6, 7 and 20.
With Aunty Stella, there were segments on different chromosomes, sometimes on the same chromosome but in different parts of the chromosome that did not overlap.

Only one overlapping segment which is on ch 7.
Then with Lena, the daughter of my mom’s half-brother George.

Lena also shared different segments and different chromosomes with Raychelle that my mom does not have, except for ch 7.
So what is consistent with all of them is that a segment on chromosome 7 is shared with Raychelle.

The diagram above shows how everyone matches each other, with the last one again showing my mom with Raychelle and that consistent block of segment.
So the fact that we all shared an overlapping segment in common with each other indicates that particular segment was inherited from our common ancestor. In this case, both Isaiah LEWIS and Polly HOLMES. But how do we figure out if that segment came from Isaiah vs. Polly? Remember that there was a discrepancy that Polly’s two husbands – Isaiah LEWIS and John LEWIS were paternal half-brothers according to some other genealogy and that Isaac LEWIS KANAE was Isaiah’s son, while John George LEWIS was John LEWIS’ son. Both Isaac and John had the same mother – Polly HOLMES.
The best way to distinguish that inherited segment being inherited from Isaiah LEWIS or Polly HOLMES is to test members of each of those families. That would be distant relatives of whom we cannot find a connection to just yet. Instead, I used another method.
Since my mother tested at 23andme, they have the ability to show the ancestry broken down by each chromosome. This is what my mother’s 7th chromosome looks like.

23andme identifies portions of the Hawaiian segments of the chromosome as a combination of East Asian & Native American, and Oceanian. I simplified it by just indicating Hawaiian. Both of my mother’s parents were Hawaiian, but her mother Rose KANAE also had European ancestry. Which is why in that diagram one chromosome is labeled as the paternal chromosome, the other as the maternal.
My mother’s maternal grandmother was Rose HOLBRON. Rose’s paternal grandfather John HALBORN was from Hull, England, and her maternal grandfather William LUDLUM was an American whaler from Jamaica, Queens, New York. Rose HOLBRON’s grandmothers were Hawaiian (Kanaka).
But it is Rose KANAE’S father – Frank KANAE whose paternal grandmother Mary LEWIS KANAE’s father was Isaac LEWIS KANAE. Isaac’s father was Captain Isaiah LEWIS. Isaac’s mother Polly HOLMES was the daughter of Oliver Holmes of Kingston, Plymouth, Massachusetts and Mahi, daughter of the chief Kalanihooulumokuikekai of Ko’olau. My assumption was that the European portion from Rose KANAE’s father is too far back. In other words, the European portion of that chromosome that my mother inherited from her mother could have only come from John HALBORN or William LUDLUM, or a combination of both.
There are a few factors that could make a segment remain in tact for several generations:
1) The length of the chromosome.
2) How many cross-over events there were for that particular chromosome.
3) Location on the chromosome (some areas are more SNP dense than others).
4) The possibility of having fewer cross-over events or none at all (we see this happening as well).
This segment seems to match nicely ranging from 7.1cM (my mom) to 9.1cM (Aunty Stella) with all the relatives.
So when I visually compare the section of chromosome 7 that matches up with the shared overlapping segment for all of us, this is where they line up.

If you have read my other posts, you would have read that multiple segments for Polynesians can remain for awhile given that we come from a few common ancestors multiple times. This paritcular segment had to have come via Polly HOLMES’ mother – Mahi who got it from her parents Kalanihooulumokuikekai and his wife. And since Raychelle is also a descendant of Polly HOLMES and Isaiah LEWIS, this portion of chromosome 7 did not come from my HOLBRON side.
While my family members used for comparison descend from Isaac LEWIS KANAE’s daughter Mary LEWIS KANAE, there are other descendants through Mary’s sister Papanaha LEWIS KANAE who got DNA tested. But only one of them was a match to Raychelle.

This cousin shares an overlapping segment of 8cM on chromosome #7. But when I compared that relative to my mother, they did not share that particular overlapping segment, although all my other close relatives did share that overlapping segment with this cousin. After looking into it further, I found out that my mother seemed to have inherited a smaller section of that overlapping segment compared to other family members, and her matching criteria just did not qualify as a match according to GEDmatch.com where all of this analysis was done. After all, she shares the least out of all the relatives only 7.1cM of this segment and Aunty Stella shares 9.1cM. And while she gave me and my brother Kaimi this segment, my brother Travis did not inherit this segment. Which means this portion of chromosome 7 for him was from our grandfather, not our grandmother Rose KANAE.
But that is what is complicated about mapping out segments for Polynesians. These segments could be from any of these lines going back to the same common ancestor multiple times. That means that Raychelle could just so happen match all of us via my maternal grandmother Rose KANAE’s mother’s side, or my great-grandfather Frank KANAE’s mother’s side, or John KANAE’s father’s side, and so forth. It could also be just by chance, that we share the segment with any other of her Hawaiian ancestors.
Since many Polynesians share multiple small segments and as small as 7cM, as well as having these segments line up very close to each other if not right next to each other, it makes chromosome mapping very difficult to do. For example, I mentioned one of Papanaha LEWIS KANAE’s descendants share that same overlapping segment on chromosome 7 with the rest of us, while the other descendants share multiple non-overlapping segments. I cannot easily assign them to our common ancestor – Isaac LEWIS KANAE, or presume that all of these multiple segments came from our common ancestor.
Since Polly HOLMES is 6 generations away from my mom and all of her descendants share this same overlapping segment, it is safe to presume that this segment came from Polly HOLMES’ mother – Mahi. And now I can assign at least this small portion to Mahi.