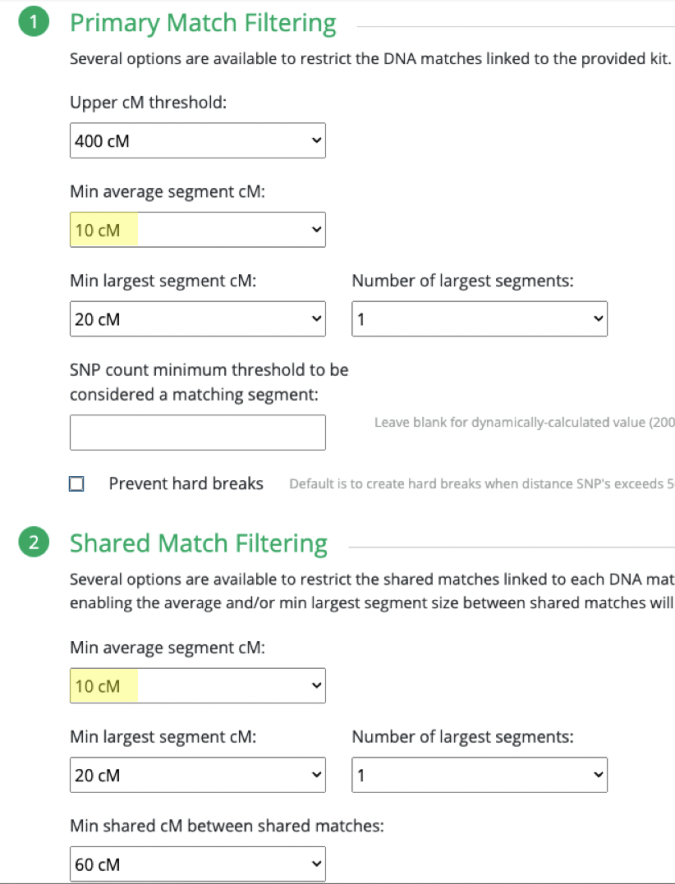
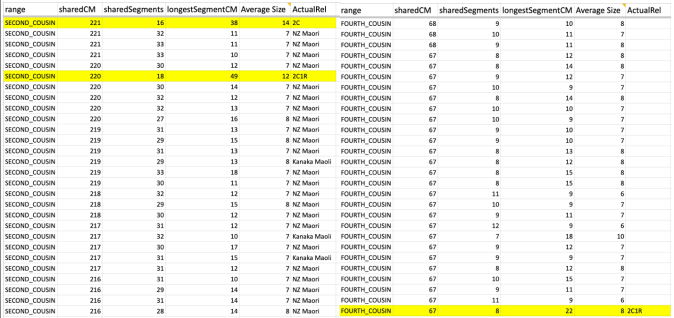
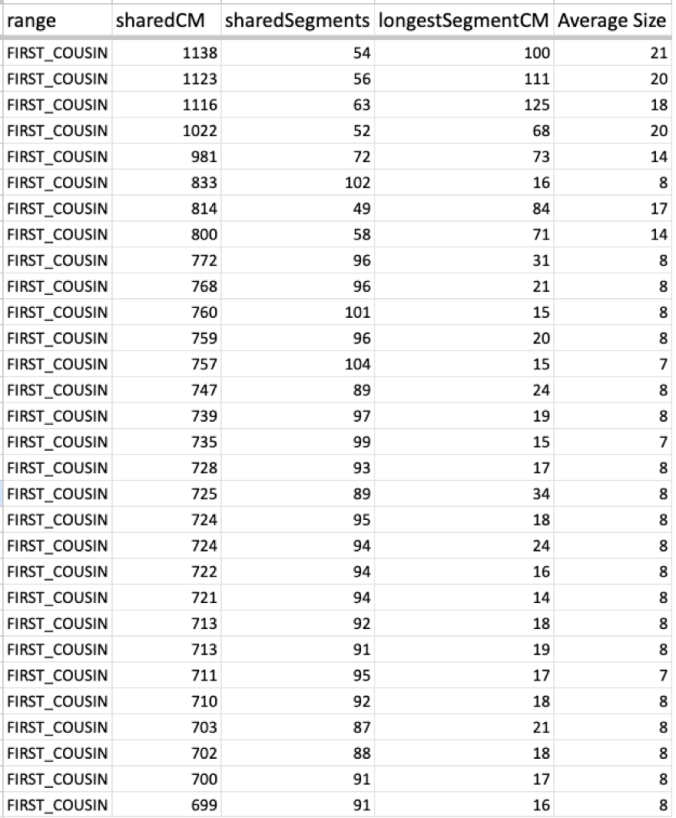
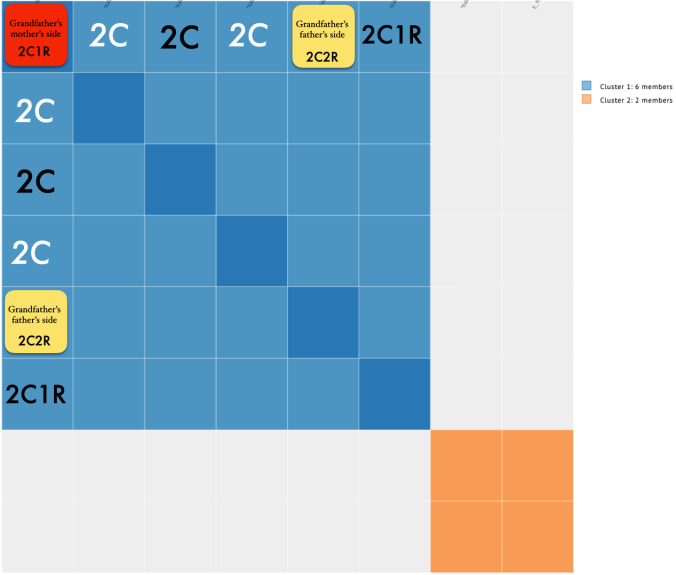
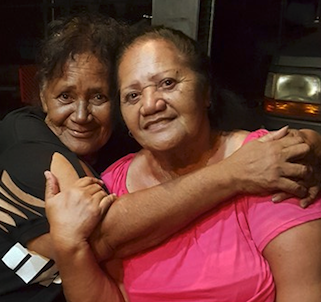MISLEADING CLOSE MATCHES
Shortly after getting my DNA results back in May 2013, I learned that majority of my DNA predicted connections are an endogamous connection. That means a predicted connection that appears to be much closer than it really is. Being Polynesian (Hawaiian), I am a result of generations of constant bottlenecking and founder’s effect that have occurred through the centuries. This effect is much more pronounced among eastern Polynesians like Maoris and Hawaiians whose homeland were the last places in Polynesia to be settled.
Since my mother was adopted and both of her parents were Hawaiian, I knew it was going to be a bigger challenge. Like other Polynesians, documentation for genealogical purpose was limited and it was not until 1860 when King Kamehameha IV passed an act to regulate names did surnames begin appearing for Hawaiians. Even right after that, surnames appearing within families were inconsistent and it varied between families, generations (some starting it later than others) and also islands.
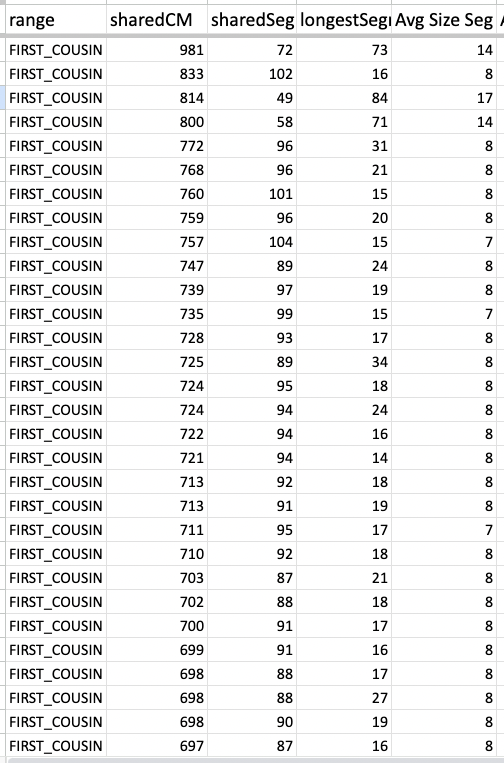
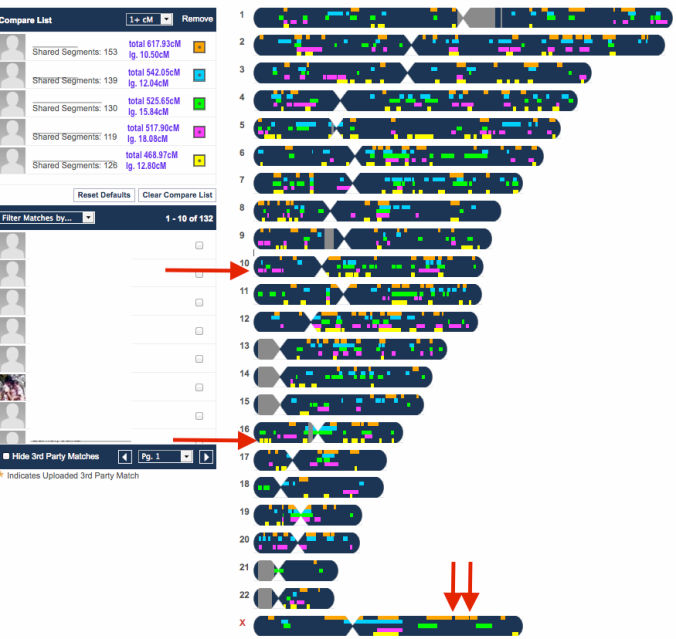
At FTDNA, my mother’s matches can have a total shared cM way above 300 (5 pages of those), while her longest block [largest segment] size tends to stay under 20cM. These are the matches from her 1st page.

FTDNA’s requirements for a match allows the tiniest segments to be included once the criteria of the longest block has been met. But in an endogamous population, specifically Polynesians, they tend to report the number of segments to be well over a hundred.
23andme is slightly different. At the default, the matches are sorted by relationship which is shown on the left column, while on the right column is sorted by percentage.

This is how GEDmatch.com shows the matches.

I expected the matches at Ancestry to be not as close as they were predicted for the same reason seen with FTDNA, 23andme and GEDmatch.
1) Several matches totaled above 100cM.
2) The largest segment usually does not exceed 20cM.
3) Matches are usually Hawaiians who do not match each other at the same amounts, and Maoris of whom we share a distant connection from 800 years ago.
As I looked at the Ancestry matches and compared the predicted 2nd – 3rd, or 3rd – 4th cousin predictions and how they appeared on GEDmatch, I noticed that they would fall in a large range of 187cM – 304cM for the total autosomal shared.
At the time I was trying to compare this close relative, Ancestry did not have the option to see how much you share, unlike now where they list the total amount of centimorgans you share and the number of segments. But still no chromosome browser to do a full comparison. Being able to see the largest segment would be key in determining a true close or distant 2nd or even 3rd cousin relationship.

So there are four matches in the 1st – 2nd cousin range (Extremely High) but I could not determine if they just appear to be close, or were true 1st to 2nd cousin matches. Given the pattern with the other companies and GEDmatch, I ignored Ancestry for over a year, until earlier this month when I reached out to my mother’s best match at FTDNA.
DETERMINING THE BEST MATCHES FOR TRIANGULATION
My mother’s best match belongs to a woman who tested at FTDNA. They share 266.94cM (122.9cM GEDmatch) total, and the largest segment is 50cM. A largest segment size of 50cM is a very good indicator that the relationship was not too distant. I guessed somewhere around a 2nd to 3rd cousin.
Last year she shared her tree with me that goes back to her ancestress named Theresa Manner, the daughter of a Swiss man and a Hawaiian woman named Kama’u. This match’s paternal grandmother and paternal grandfather were both Hawaiian. Kama’u was an ancestress on the paternal grandfather’s side.
Back in January I began focusing on Theresa Manner’s husband’s line, especially since his family lived in the area where my mother was born. Not to mention I had nothing else for Theresa Manner except her parents’ names, and Kama’u was the only Hawaiian that I saw in that line.
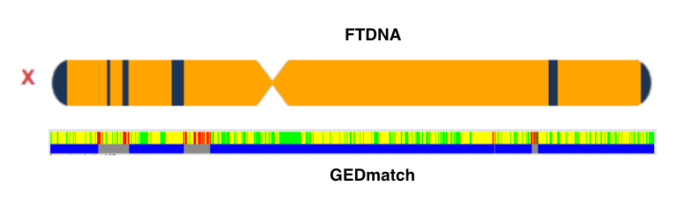
My match asked me if I thought there was a possibility that it was her paternal grandmother’s side versus her paternal grandfather’s side. Her paternal grandmother was also part Portuguese, but since we were not getting matches with Portuguese people I excluded the paternal grandmother’s side. Although this match and my mother share 2 segments on the X chromosome, the largest segment 10.2cM while the other 8.8cM, I knew it had to be distant given the unpredictability of the X particularly for Polynesians. So I ignored it and continued to focus on the paternal grandfather’s side.
At Ancestry, my mother’s closest match belongs to a man and is the first match at the top of the diagram above. But this match’s tree did not have any names in common with any of the other top matches that we get. No matter how many conversations I have had with this closest match at Ancestry, although all prior to my mother revealing to me in August 2014 that she was adopted, I still was unable to find any close connection. Given the endogamous history and the fact that we just lack genetic diversity, it seemed more of a validation that the match was not as close as it appeared to be.
ANCESTRY RESULTS TO HELP NARROW DOWN SPECIFIC ANCESTORS
I also focused on my mother’s ancestry, which points to two basic ancestries. East Asian and European. Her Polynesian portion is usually represented by the East Asian and Oceanian categories combined. Some companies such as FTDNA’s “myOrigins1” have lumped Oceanian under their East Asia category which previously their “Population Finder” separated them. 23andme also separates the two categories while AncestryDNA recently created a category called Pacific Islander (Polynesia). Below are the results from those companies including analysis from Dr. Doug McDonald2. For simplification I combined the Oceanian with East Asian.

AVERAGE
81.72% = East Asian/Oceanian
17.32% = European
This next diagram shows 23andme’s chromosome view. My mother’s X chromosome was just East Asian/Oceanian in origin. Dr. Doug McDonald also had a chromosome view and he too found that it was only of East Asian.

Knowing the X inheritance pattern, plus the amount of European percentage that my mother has, I tried to calculate who would have been the most likely European ancestor that married a Hawaiian. If not European, then of European descent. So I had a few clues that helped me determine how to figure out who that was.
1) My mother recalls meeting her biological father at the age of 5, and claims that he was pure Hawaiian.
2) Our mtDNA haplogroup is B4a1a1a33, a subclade of the Polynesian motif B4a1a1, indicating our direct maternal line as Hawaiian.
3) The odd percentages is probably the result of more than one ancestor being of European and Hawaiian ancestries.
Given those details, I constructed this diagram which would be the likely scenario of how my mother got her European and Hawaiian ancestries.

I thought that looking for her European ancestors would be easier to trace given the few early European (or American of European descent) arrivals in the Hawaiian Kingdom. Unlike with Hawaiians whose DNA results produce closer predicted relationships than they really are.
I calculated both 18 years per generation in that diagram and 25 years for each generation. The 25 year estimate took me to 1868 from my mother’s birth year of 1943, and back to a 50% European/Hawaiian person in that diagram. Theresa Manner was born in 1866, so now I was confident that this could be very useful in tracing my earliest Hawaiian/European-descent ancestor.
Comparing this diagram to Theresa Manner and knowing that the estimates of the dates were very close, I realized that I did not go back far enough. Although the predicted 50cM largest segment for a 2nd to 3rd cousin was good, it was best to be sure to go even further.
TRIANGULATION & FURTHER RESEARCH
My match at FTDNA also revealed that her father’s Y-DNA results showed a European haplogroup rather than a haplogroup indicating Polynesian origins of which her father has a direct male Hawaiian line. Given this new information and not knowing where this NPE (non-paternal event) could have occurred, either with her father or her grandfather, or even further back, I immediately excluded this line. This was the same line I was previously looking into back in January, focusing on Theresa’s husband and their children and her husband’s siblings and their children. Although still, that would not tell me for sure if that was my mother’s paternal or maternal side.

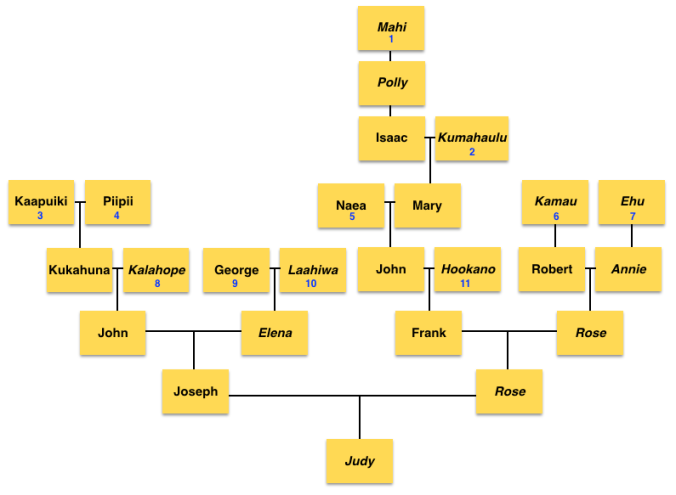
I realized how I was ignoring Theresa Manner, whose father was a Swiss and he married a Hawaiian woman named Kama’u. I asked my match if Theresa Manner had any other siblings. Previously my match only shared Theresa’s parents, and Theresa’s husband and their children. Again, because of the 50cM largest segment prediction, assuming it was pretty close, that I did not have to go back further. But I knew that Theresa Manner was an important clue since she was half European (Swiss father) and half Hawaiian and was born in 1866. That fit into the year I predicted from the diagram that I created in order to come up with the estimated 1868 birth for a male ancestor that was 50% Hawaiian and 50% European. I was told that Theresa not only had 3 other siblings, but also 2 half-siblings. They were Robert Holbron and Mereana (Mary Ann) Holbron. Kama’u was previously married to John Holbron from England.
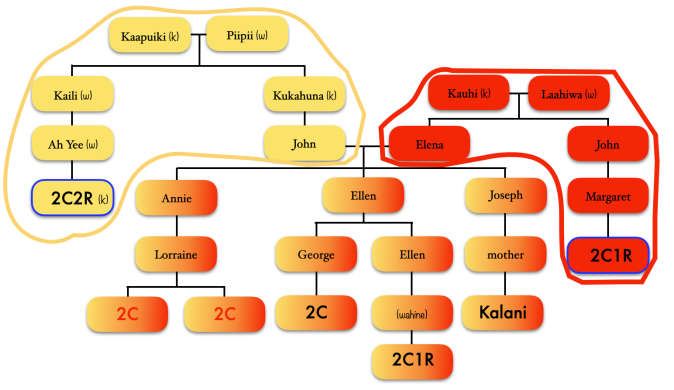
I thought that name Robert Holbron seemed familiar. Since several of our top matches at Ancestry had public trees, I went back there to look and saw that the very top match of whom I have had correspondence with last year listed in his tree Robert Holbron and his wife Annie Ludlum as his ancestors. This match is in the predicted range of 1st – 2nd cousins, although as mentioned previously, it may or may not have been a true 1st – 2nd cousin relationship. But the fact that both of these matches had Kama’u in their line and of whom was probably the one responsible for passing on that large 50cM segment, I knew I found my ancestors.
With this new information things fell into place. I referred to the diagram I created and it seemed that Robert was the more likely candidate to fit into what I had constructed as my possible ancestors. But to be sure, I looked into his sister’s descendants first. Mereana Holbron married an Irish man, lived in Ireland and in Hawaii. But Robert appeared to fit the pattern I mapped out more than his sister, although his wife Annie had a surname of Ludlum, which indicated that she was an admixed Hawaiian unlike in my diagram. But even with her being half, I knew that her mother was Hawaiian based on the fact that if Annie were my ancestor, her mother would have passed on my B4a1a1a3 directly to Annie and down to females. And that matched up with my diagram. My match’s tree at Ancestry listed Annie’s parents as William Ludlum and Ehu, which confirmed that Annie was half Hawaiian and of half European descent. Later, I would find that both Robert and Annie’s estimated year of birth fit the diagram and predicted year of birth only off by 13 years.
While going through several of the old Hawaiian newspapers I found an article with a photo of Mrs. Annie Holbron celebrating her 100th birthday.

Annie Ludlum
SPECIFIC BRANCH NARROWED DOWN
I continued looking into Robert and Annie’s children and grandchildren. I focused on their oldest daughter Rose Holbron and her husband Frank Kanae, and who our top match at Ancestry descends from. And although I made sure I covered many of the collateral branches, I still used as a guide the diagram I created to guide me into which branch it could be. And it took me to Rose & Frank’s 3 daughters.
While looking for their descendants and who they married, one of them had a photo uploaded into their tree on Ancestry, and I saw the striking resemblance not only to my mother but also to my sister. I found information on her husband and children and she had one child born just a month before my mother was born. So it seemed unlikely that this was my mother’s biological mother. This is on the assumption that my mother’s birth date on her “legal birth certificate” is her actual birth date. I could only assume that it may have been one of the other two sisters who was my mother’s biological mother based on the fact this woman looked like both my mother and sister.
I definitely found the right family for my mother’s maternal side. It would be only a matter of time before I find out if the woman whose photo I found was my mother’s biological mother, or her mother’s sister.
EVALUATING ALL CLUES
My mother’s legal birth certificate left questionable details since I began researching 26 years ago. It was because it did not indicate that it was an adoption, unlike my own birth certificate which clearly states that it is an adoption. What was known and is clearly indicated on my mother’s birth certificate.
- Birth was not a hospital or institution but at a residence – 1301 Liliha St. (Honolulu)
- A midwife was present, attested to witnessing the birth of Julia Kawewehi Scott [adoptive mother].
- Usual residence of mother was 440 N. King St, which was the permanent residence of both parents.
I spent several years looking into the address where my mother was born. I even tried to track down people who lived at the same address hoping to find someone who was alive at the time my mother was born and may know something about her birth mother.
Now that I have found the branch that we come from, again I continued to look into Rose Holbron and Frank Kanae’s daughters. I already found one with a photo who resembles my mother and sister. Another daughter I found was married several times, and had children from possibly more than just her own husbands, but this is based on surnames. Her name was Rose Kanae, and her first husband was surnamed Kalei. Her second husband was Joseph Akana, which can be a semi-common name. A lot of Akanas, but not all are related to each other from what I saw with their last known ancestor to some of the Akanas that I found. That surname comes from a Chinese who either became a Hawaiian citizen or just Hawaiianized his name. That surname caught my attention and made me think about it for a bit, but given that I had done a lot of genealogy I know and do remember seeing that name and reading about that surname in other Hawaiian genealogy forums. Then after Joseph Akana, Rose married a third time. All of those husbands gave her children.
I was surprised to find in the city directory of 1947, that Joseph Akana lived at 1301 Liliha Street. This was the same exact address where my mother was born. So either this woman got pregnant from another man and Joseph Akana divorced her, or her sister got pregnant and gave birth at their residence.

At the same time I found the photo of the woman who could be my mother’s biological mother, my mother called to tell me that she received her non-identifying form from the First Circuit Court in Kapolei, O’ahu, Hawai’i. This form just gives adoptees little information into their background and every state and court has their own way of determining what will be revealed. For Hawaii, it is the ancestry.
When I received a copy of it, it indicated that my mother’s mother was Hawaiian and Chinese while her father was Chinese Hawaiian. However, DNA does not support that.

Non-Identifying Form – letter form
But given the non-ID form indicating Chinese, although incorrect, this was pointing to Joseph Akana and Rose Kanae as the parents listed. Joseph Akana’s surname was the only clue that made me believe he was not the biological father. I had even suspected that there may have been a question of paternity with Joseph Akana and what may have caused my mother to get adopted, hence Joseph divorcing and my grandmother remarrying.
I was also told that back then, no matter which state, particularly for unwed mothers, if the biological mother knew who was going to adopt the child, she might take on the identity of that woman and check into the hospital as that person. The woman who adopted my mother – Julia Kawewehi was Hawaiian and Chinese. So I assumed this is what happened unless Rose Kanae really did not know her ancestry. The DNA evidence indicates a near perfect 70% East Asian to 30% Oceanian component consistent of Polynesians4. Whereas those who have some Asian ancestry in addition to Polynesian, the percentage of East Asian to Oceanian would be much more significant than 70%. Therefore either the parents listed in the adoption files really are not her parents or they were just wrong with the ancestries of the parents.
THE FOREIGN ANCESTORS
After getting more details on my maternal grandmother Rose Kanae’s branches, I was able to see exactly all of the foreign men who married Hawaiian women in her line.
1) Oliver Holmes, (later Hawaiianized to Oliva Homa) an American who arrived on the island of O’ahu in the Sandwich Isles on October 8, 1793 on board the Margaret, who was in service of the chief Kalanikupule. After Kalanikupule’s death in the Battle of Nu’uanu, Holmes married Mahi, the daughter of the high chief of Ko’olau5 whose name was Kalaniulumokuikekai6. After Kalanikupule was defeated by King Kamehameha, along with Mahi’s father Kalaniulumokuikekai, Oliver and others were taken by Kamehameha and became advisors for the King. In 1810 after King Kamehameha united all the islands, Oliver became the 3rd Governor of O’ahu. Governor Homa, as he was called, remained in Honolulu and had one son and five daughters. One of the daughters – Hannah Holmes married Captain William Heath Davis Sr, and their son William Heath Davis, Jr. ended up in California in San Francisco and was also one of the original founders of New Town San Diego7. Oliver’s other daughter Polly married Captain Isaiah Lewis and it was said that she traveled with him to the northwest coast. Oliver had three other daughters, all of whom married captains of ships coming into the Hawaiian islands.
2) Captain Isaiah Lewis, an American and son-in-law of Oliver Holmes. He commanded the Panther, and acquired sandalwood in Hawaii and sold it in Canton, China. He married Polly Holmes, the daughter of Oliver Holmes & Mahi.
3) William Ludlum, an American whaler from Jamaica, Queens, New York who became a citizen of the Hawaiian Kingdom on August 7, 1850. He married a Hawaiian woman named Ehu on January 24, 1850 in Mapulehu, Molokai. He ran a hotel & was a Commission Agent.
4) John Holbron [John Halborn/Holborn], originally from Hull, England arrived in the 1840s. He married a Hawaiian woman named Kama’u and became a citizen of the Hawaiian Kingdom on November 24, 1845. He was a merchant.
After knowing for sure who these foreigners were, I was able to re-design that diagram and came up with new percentages of each ancestor.
EA/Ocean = East Asian/Oceanian
 The light blue represents the male, the pink the female, and the bottom is my mother of which is calculated 14% European and 85% East Asian/Oceanian. My mother’s results fall into that range of 14% – 19.81% European and 80.18% – 85% East Asian/Oceanian.
The light blue represents the male, the pink the female, and the bottom is my mother of which is calculated 14% European and 85% East Asian/Oceanian. My mother’s results fall into that range of 14% – 19.81% European and 80.18% – 85% East Asian/Oceanian.
MISLEADING X MATCH
While my match at FTDNA did share 2 X segments with my mother (via her father of whom both of his parents were Hawaiian), the fact that I ignored it because of how unpredictable the X is lead me on the right path. The largest segment size is 10.2cM and the other is 8.8cM. I have seen how a significant amount of X my mother shares with other Polynesians (i.e. Samoans, Tongans & Maoris) can be so distant.
While it can be useful in excluding certain branches, this is obviously not the case for endogamous groups, particularly not for Polynesians. Had I focused on the X path, I would not have made the connection at all. But it is not surprising that this male shares a distant X match with my mother on his mother’s side yet not have a recent tie to my mother on his maternal side, at least not that I have figured out just yet.
CONFIRMATION OF BIOLOGICAL PARENTS
My friend Charles Ano who have helped me with genealogy searches in the past decided to look up birth announcements in the newspaper around the time that my mother was born based on all of this information that I found.

Birth Announcement – Honolulu Advertiser – Sept 28, 1943
It listed her original name – Joyce Awapuhiokala (Akana). I did not want to reveal this to my mother just yet since I completed the last step of accessing her adoption records just the other week. I wanted my mother to receive copies of the actual adoption files and see for herself who they listed as her parents and her original name.
Eventually my mother found out that I knew of her original name, and after I got a chance to speak to her about it, she told me that she already knew she was born Joyce and that she was related to the Akana family, and also had a Hawaiian name, but she did not know what that name was. She was about 9 years old when she found out about all of this.
I did tell her that I remember her mentioning the surname – Akana. And I knew that when I found Joseph Akana, there was something about that name that seemed familiar but could not remember exactly. I later realized it was last year after almost giving up on DNA and before resorting to accessing her adoption files via the court, I had turned to the 1940 census, just three years before she was born and looked for the place where she was born. It was at that time I asked her about the family names, people who lived at that address where she was born to see if it seemed familiar. None did, but it was at that time when she asked, “what about Akana?” I never asked her much more about it, but remember she did comment about how she heard that was her family. Unfortunately, I forgot about this important clue.
CONTACTING COUSINS – BIOLOGICAL FATHER CONFIRMED
Now knowing my mother’s original name, I decided to seek out my cousins, children of my mother’s siblings. I already saw names of some of them based on the obituaries I found of my mother’s siblings. I first contacted a woman whose surname matched my mother’s brother and with whom I already had contact with because at the beginning of accessing my mother’s adoption files I simultaneously tried to get my original birth certificate since my adoption was done at another court. My adoption was a technical issue. The mother that I speak of is my biological mother. It turned out that this contact is a granddaughter of my mother’s brother. After sharing with her who my mother is, what name she was born, who her parents are and identifying the other siblings based on what I found in the obituaries, she notified both her grandfather and a sister living on another island. I sent photos of us and she shared it with her great-aunt and grandfather. She remarked at the strong resemblance of my mother and her great-aunt.
At the same time, I found another cousin on Facebook, contacted our mutual friend and that friend revealed how this cousin probably knew my mother. Turns out that this cousin and my mother have known each other for the past 6 years. This cousin also told my mother at first meeting her 6 years ago that she reminded her of his aunt. This was the same person that the other woman commented of how my mother looked like her great-aunt.
In a few days after having other cousins contacting me, they revealed the entire story of how our grandmother Rose Kanae married three times, and with her husband Joseph Akana she had four children. A daughter who carried the surname KALEI which was the surname of Rose’s first husband. Another daughter and of whom is the only sister to my mother that is alive, the one who we are told my mother looks like. Then a brother and finally my mother. Then with the last husband, Rose had two sons. The youngest is alive and whose granddaughter I first contacted.
Only the sister that is still alive and one of the younger half-brothers were the only two children that were not given up for adoption. The other seven children were all given up for adoption. The older ones knew of my mother’s existence. But what they knew was that they had a sister named Joyce Akana who was given up for adoption by a Filipino family. My mother’s adoptive father was Filipino.
Then it was revealed to me how Joseph Akana really was my mother’s father, but Akana is a surname he took on later in life and was the surname of his paternal aunt’s husband. His original name was Joseph Napua Kaapuiki and was a pure Hawaiian man just as my mother remembered. And Joseph did frequent the area and since my mother grew up right by where she was born, it is no surprise that she actually did encounter her father.
And as difficult as this was given the situation with Polynesians and endogamy, it can still be done. It would just mean that every match, especially when it comes to geographic location should be scrutinized in order to determine a really close match or not.
I was there on November 1, 2015 when my mother and her sister Stella got to meet each other.
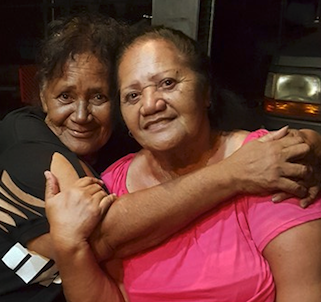
My mother Judy on the left, with Aunty Stella on the right.
This blog entry was edited after new evidence confirmed my mother’s paternity and after she was reunited with her sister. In May 2016 my mother received her adoption files which listed her biological parents as Joseph Akana and Rose Kanae, listing my mother’s name as Joyce Awapuhiokala Akana aka Joyce Awapuhiokala Kanae.


1. MyOrigins v1 May 2014 – April 2017. MyOrigins v2 separated the Oceanian from the East Asian and currently Oceanian falls under the broader Central/South Asia category.
2. Dr. Doug McDonald developed a the Biogeographical Analysis software and was contracted with Family Tree DNA to write the underlying code for their Population Finder ethnicity software.
3. Phylotree build 17 currently is B4a1a1c.
4. Population Genetic Structure and Origins of Native Hawaiians in the Multiethnic Cohort Study shows that autosomal results for Native Hawaiians consist of 68% Southeast Asian and 32% Melanesian components.
5. Day, A. Grove. (1984) History Makers of Hawaii. Honolulu: Mutual Publishing of Honolulu. pp. 53.
6. Honolulu Star-Bulletin – Monday, June 20, 1960: Tales About Hawaii – Oliver Holmes Founds a First Family.
7. William Health Davis, Jr.