In my last entry I demonstrated the difficulties of determining the half-relationships after receiving the DNA results of my half-first cousin. Within an endogamous group, that could be even more difficult as we see larger amounts of DNA shared.
While the ISOGG Wiki Autosomal DNA Statistic page can list the average amount of centimorgans shared, Blaine Bettinger’s The Shared cM Project demonstrated that the minimum and maximum amounts shared can vary. This becomes more evident as the distance of relationship increases.
Within an endogamous group it makes sense that having more than one pair of common ancestors may increase that amount. The same would apply if you descend from the same common ancestor multiple times. Both would produce higher amounts shared.
A few months ago I got the results of my aunt believed to be a full-sister of my mother. My aunt suspected that her father was not her biological father. And she was right. But she was not the only one who knew of this, but the rest of the family, particularly the ones of my generations believed that this Aunt’s father was her biological father and did not suspect otherwise.
From my mother’s Family Finder (autosomal) match list at FTDNA:
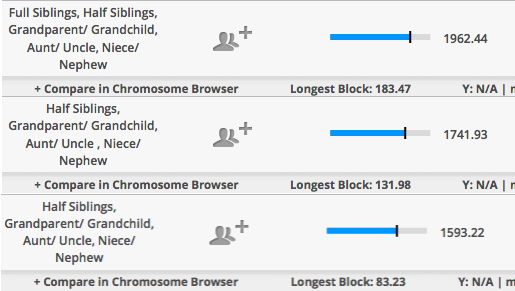
The top is my mother’s sister while the one right below it belongs to my half-1st cousin whose father George was mentioned in the last entry – Determining half-relationships with Polynesians.
Initially I was confused by the total amount since I knew it was more than what I shared with two of my half-brothers. This is how two of my half-brothers compare to me and to each other.
So my mother and her sister did share a bit on the high-end for half-siblings, but low end for full-siblings. These are the predicted averages shared for siblings vs. half-siblings.
The next step was to take a look at the X chromosome. For half-sisters who had the same father, they would share an entire X chromosome based on how the X is inherited. To my surprise, it looked like someone took a razor blade and sliced out some pieces of the image.
For half-sisters they share a lot compared to what I saw when comparing my half-brothers to each and to me. Also, I decided to include both the default 5+cM setting and the 1+cM. With my brothers, we hardly get anything when I lower it to 1+cM. But with my mother and aunt, you can see a difference although chromosomes 4 and 18 are more likely to be IBS, but given the situation (endogamy, small communities, & isolation) it just may be IBD from a very long time ago.
So the X was not helping me one bit since I thought maybe they were areas on the chromosome that could not be read – no calls.
I immediately uploaded to GEDmatch for further analysis. No surprise that when I looked at the X, it was the same exact thing. Knowing that it wouldn’t be helpful, I turned to the other 22 pairs of chromosomes.
What you would be looking for in full-siblings are full-identical regions (FIR) which are the green sections on the bar graph. Here is an example of my 1st cousins, a brother and sister.
About 25% will be fully identical. You can read more about how much full versus half-identical regions siblings would share at ISOGG’s Wiki – Fully Identical Region page.
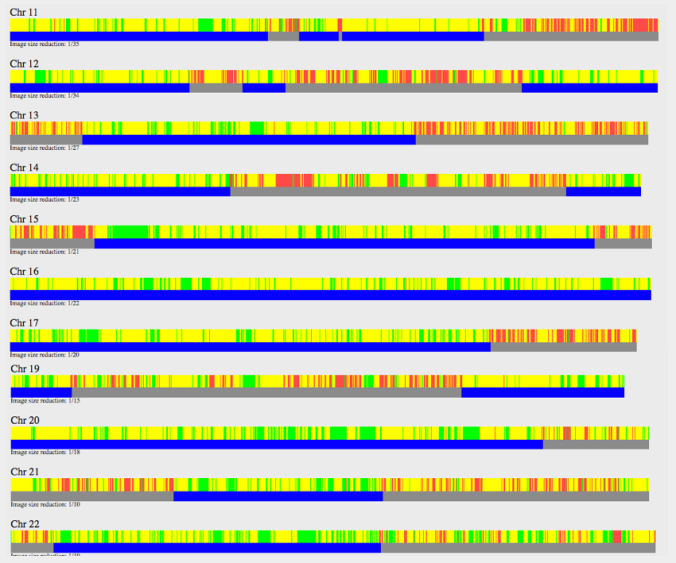
This is what my mother and aunt showed.
There are only small chunks of FIR rather than long segments of it that you would see in full-siblings. So this confirms a half-sibling relationship.